Bolivian biologists building a better future for science and health
Three generations of scientists visit CZ Biohub SF to learn metagenomic sequencing techniques and analysis
For the past century, diagnosing malaria infections in the developing countries where the disease is endemic has been a tedious and imperfect process. Armed with a field backpack, dedicated technicians perform pinpricks on patients and use microscopes to analyze slides containing thousands of stained blood cells, of which only a few may contain the Plasmodium parasites that cause malaria.

Microscopist Sokunthea Sreng, of the National Center for Parasitology, Entomology and Malaria Control in Cambodia, analyzes blood samples in the field. Photo: Jessica Manning
The so-called Giemsa stain favored for this task, first devised in 1902, can be quite a sensitive test when used with the thick smears of blood obtained through pinpricks, since there are many blood cells in the field of view and a good technician can pick out parasites as different colors pop in the images. But it is a notoriously difficult and finicky protocol: platelets and other blood cells also light up with Giemsa staining, and even changes in temperature can affect results.
“People who use Giemsa stains can rightfully claim that they have the ability to detect extraordinarily low amounts of parasites on thick blood smears,” says Paul Lebel, an R&D engineer with the Chan Zuckerberg Biohub’s Bioengineering Platform, led by Rafael Gómez-Sjöberg. “But what they’re leaving out is the fact that, without a gold standard to compare against, they’re getting it wrong half the time.”
Now, thanks to advances in the field of deep-learning AI, and the availability of cheaper microscope parts, Lebel and a group of CZ Biohub colleagues have created a blueprint for a device with the potential to replace Giemsa staining as the go-to malaria diagnostic in low-resource settings—an accomplishment that “would not have been possible 10 years ago,” says team member and CZ Biohub Co-President Joe DeRisi.
The microscope prototype is based on findings published by the Biohub team on August 9, 2021 in the open-access journal PLOS Computational Biology. The work began with the desire to develop a diagnostic that doesn’t use any label or stain—these rely on expensive reagents and require cells to be killed by fixation. Moreover, “there’s a lot of bad Giemsa staining that happens during these steps, so you waste blood and you waste time,” says Lebel.
Biohub Data Scientist Pranathi Vemuri joined Gómez-Sjöberg, Lebel, and DeRisi in the new work, along with Rebekah Dial, now a research scientist at Denali Therapeutics, and Valentina Garcia, a graduate student in DeRisi’s UC San Francisco lab. Biohub Senior Engineer Chris Charlton is also part of the team building the prototype.
Previous attempts at label-free imaging of Plasmodium, for instance with a technique called quantitative phase microscopy, have not only required expensive, specialized optics, but they have failed to detect the parasite in its predominant form—the immature “ring” stage. But “ordinary images contain all the information we need,” Lebel says. “The key is very high-quality annotated data and rigorous validation. The classifiers will learn to extract the right patterns from those.”
In the work reported in their paper, the team used live red blood cells as a proof of principle before they move to using whole blood, and found that microscope setups that illuminate the sample with very short wavelengths of light, from deep UV to barely visible 405 nm, allow the best combination of high resolution and contrast to resolve the ring form as well as the trophozoite and schizont forms of the parasite in infected cells—without using any stains.
Peering through these microscopes for many long hours, Lebel became adept at spotting the different parasite stages within their lab-infected cells using either a custom-built deep-UV microscope or a standard commercial device customized to have brightfield illumination at 405 nm.
But to turn these findings into a reagent- and time-sparing tool for field scientists’ backpacks, the team turned to deep learning to automate the identification steps.
The group didn’t have to build any new deep-learning networks, as existing ones could be retrained with manually sorted images of defined parasite stages they had obtained. The innovation came in the form of extensive image annotation and validation, and ultimately the resulting automated classifier performed better than the gold standard, manually annotated Giemsa staining. The validation step included a comparison to a “ground truth” sample—a Giemsa-stained blood sample with a very high parasite load—and the new system performed with extremely high accuracy.
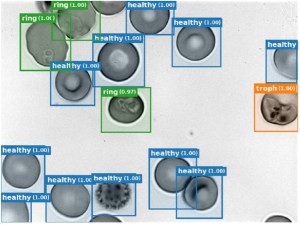
The automated classifier in CZ Biohub’s prototype malaria diagnostic device rapidly detects individual blood cells and annotates them as either uninfected (“healthy”), ring, trophozoite (troph), or schizont. Courtesy of Paul Lebel.
The result? A microscope that can automatically and rapidly distinguish infected red blood cells from healthy ones, and can classify infected cells by stages of the Plasmodium life cycle.
In Cambodia malaria is endemic and virtually all diagnoses are currently done with the Giemsa technique. The team hopes that by year’s end their first low-cost, field-ready prototype will be deployed there for testing, and it should be able to process on the order of a few hundred thousand cells in a 20-minute interval, as opposed to the few thousand a human might look at using Giemsa staining.
Besides saving labor, the new Biohub device will surely improve diagnostic accuracy in the field. According to a competence scale for skilled Giesma-stain technicians outlined by the World Health Organization, the most accurate readings will be within 25% of the correct answer only 50% of the time. “Without the ground truth,” says Lebel, “it’s basically a toss of the die.”
Three generations of scientists visit CZ Biohub SF to learn metagenomic sequencing techniques and analysis
Learn More
A ‘Google Earth’ of embryology, CZ Biohub San Francisco's zebrafish cell atlas brings a new vision to developmental biology
Learn More
A rock-climbing scientist brings his exploratory spirit to science and medicine
Learn More
Stay up-to-date on the latest news, publications, competitions, and stories from CZ Biohub.
Cookies and JavaScript are required to access this form.