The future is now: four insights from CZ Biohub Chicago’s inaugural scientific conference
Researchers gathered to discuss trends and technologies that will enable biological research
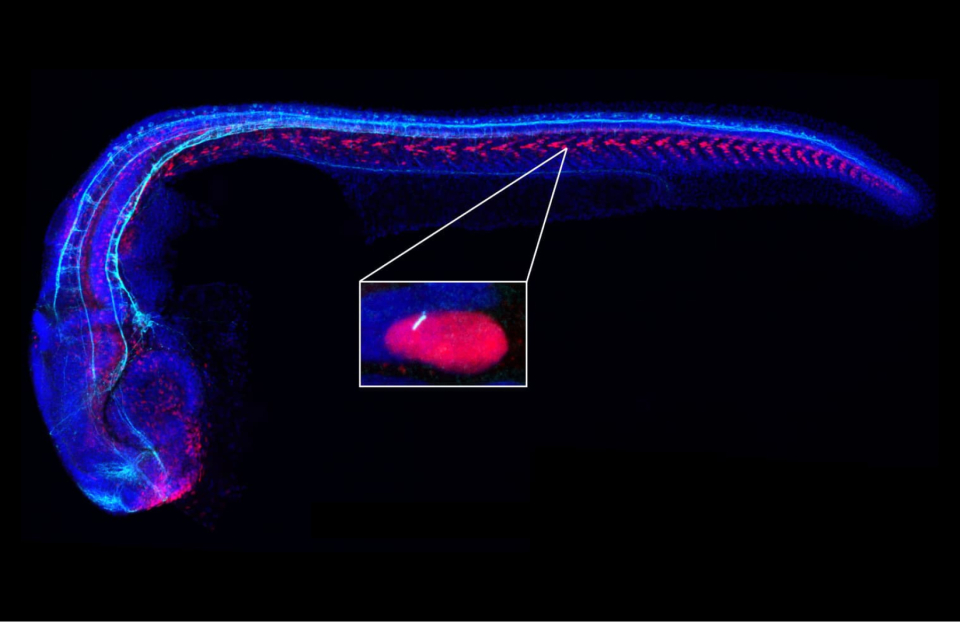
In the inset, a primary cilium (white) is seen protruding from a cell in a zebrafish embryo. (Courtesy of J. Reiter, UCSF)
Nearly all vertebrate cells are ciliated, with slim appendages that can make the cell motile or, in the case of non-motile or “primary” cilia, serve to compartmentalize signaling pathways into a very small space. Primary cilia represent just a fraction—1/3000th—of the cell’s volume, and were once considered vestigial, but they are now known to contain hundreds of different proteins, some of which, compared to the cell body proper, are highly enriched in these antenna-like structures.
Among these cilia-concentrated proteins are more than 30 membrane-embedded G protein-coupled receptors (GPCRs) responsible for receiving specific signals, so-called ligands, from outside the cell, making primary cilia a sizable signaling platform—with a quite high surface-to-volume ratio—and handy cell-biological arrangement. And because each cell’s single cilium literally sticks out from the surface of the cell, they are well-positioned to receive signals coming from the external environment, and also to contain any machinery required to perform the computation required to decide how to respond. However, formal evidence of the cilium as a signaling sub-compartment has been sparse.
Activation of GPCRs triggers production of the small molecule cyclic adenosine monophosphate (cAMP), which amplifies the receptor’s signal and moves the action from the cellular membrane towards the interior of the cell and ultimately to the nucleus, where the business end of signaling takes place. This poses an immediate problem, since cAMP is also being produced downstream of various GPCRs, and these GPCRs are present on every surface of the cell (cilia and otherwise). The decision of which downstream pathway to turn on could be easily overwhelmed and confused.
Subcellular compartmentalization—a defining feature of cell physiology—could offer a solution. In classic work, Edwin Krebs and Joseph Beavo and others suggested that one of cAMP’s three effectors, protein kinase A (PKA), as well as its effects, are spatially localized—different ligand-receptor pairs use different micro- and nano-domains of the membrane.
Likewise, subcellular organelles, including cilia, could serve a similar function to organize cAMP alongside its appropriate effectors within the cell. Several signaling pathways occur exclusively in primary cilia, including Hedgehog (HH) signaling, a pathway that is fundamentally important in various developmental processes. Within primary cilia, HH binds its receptor to activate the GPCR-related Smoothened (SMO) upstream of Gli transcription factors that have to find their way to the nucleus to be able to transmit the signals that lead to the ligand-specific cellular changes.
Chan Zuckerberg Biohub Investigator Jeremy Reiter and members of his lab at UC San Francisco have been interested in the question of spatial regulation of signaling and other puzzles surrounding cilia for some time, including from a translational perspective: because of their critical roles in cellular and organismal physiology, dysfunctions in various cilium-associated structures and processes can lead to a number of diseases, called “ciliopathies.”
In new work published April 30, 2021 in Cell, Reiter’s team, including former graduate student and first author Melissa Truong, worked with the lab of another CZ Biohub Investigator, UC Berkeley’s Ke Xu, to show that cAMP generated in cilia is interpreted differently than cAMP produced in the cell body, allowing for unique responses to the same signaling mediator—the same molecule has different effects based on its localization.
To arrive at this intriguing finding, the Reiter lab employed a series of elegant optogenetic and chemogenetic tools to manipulate cAMP distribution and GPCR signaling as well as a biosensor that measures relative cAMP levels. Optogenetic and chemogenetic tools have been widely used over the past 15 years to make a protein’s function light-responsive, but have been mostly applied in single cells. Here, the authors applied them in whole living zebrafish embryos.
They also did similar tricks to localize PKA to the cell body; to the base of the cilium (the basal body); or to the interior of the cilium itself, all within the intact embryos. When they used their tool, called bPAC, to generate cAMP selectively in either the cilia or the cell cytoplasm, they found that cAMP could only reduce HH signaling if is generated in the cilium, not in the cytoplasm. They saw a similar phenomenon in the well-studied tissue-culture cell line NIH/3T3.
Armed with the finding that cAMP can convey different information to initiate distinct downstream effects based on where it is generated, Reiter’s team now had to explain how the highly diffusible cAMP could be contained within the cilium and not find its way to the cell body to impinge on pathways unrelated to the original signal.
The Xu group has long explored the interplay between experiment and theory, using microscopy data to inform in silico models of how biomolecules move within their native environments. Xu’s curiosity was piqued when he heard Reiter give a presentation at a CZ Biohub Investigator meeting which posed the question of how cells could impart a unique response to HH-stimulated cAMP versus the many other sources of cAMP.
To approach Reiter’s problem, Xu’s group built a computational model of simple diffusion that also takes into account cAMP bouncing off the membrane. The approach employed a spherical model cell in which PKA is distributed according to microscopy data; where collisions with cAMP activate PKA; and where there is no barrier separating the cilium and the cell body, so cAMP can freely move about.
Using this model, the collaborators were able to conclude that the approximately 13-fold greater surface-to-volume ratio of the cilium is sufficient to explain how PKA in cilia is activated more quickly than that in the cell body. In addition, the same amount of cAMP in the cilium or cell body may result in a much higher local concentration of cAMP available for activating PKA in the cilium.
With this geometric explanation for how different subcellular locations can impart different information to the cell, even if these locations are contiguous and the signals are diffusible, Reiter’s group went on to show that ciliary cAMP acts through ciliary PKA, but doesn’t affect PKA in the cell body. The bottom line? PKA in the cytoplasm is functionally distinct from that in the cilium.
UCSF’s Wallace Marshall, part of a CZ Biohub Intercampus Research Team exploring the biology of atypical model organisms, notes the distinction between compartmentalization in cilia versus other cellular organelles. The latter are generally spherical, so their surface-to-volume ratio drops as they grow, while in cilia that ratio stays the same with a change in size. The cell as a whole can therefore vary its response to signals, much like a tunable circuit, depending on the geometry of the compartment where the receptors are localized, just as the Reiter and Xu work found in cilia. But cilia are not a special case, as organelles and even phase-separated condensates provide unique signaling platforms, which also happen to be located in the interior of the cell.
Indeed, to complete the cAMP-driven signaling circuit, we necessarily must look to other cellular compartments, including the nucleus. Mark von Zastrow, also leading a research team at UCSF, has been studying cAMP signaling downstream of various organelles. He has been inspired by classic work from Eric Kandel’s group showing that cAMP signals initiated at neuronal synapses resulted in transcriptional changes in the nucleus that are involved in long-term plasticity that contributes to forming and consolidating memories.
With respect to the Reiter and Xu work, von Zastrow notes that compartmentalization to the cilium is the first part of the answer to the question of how cAMP signals can travel across those vast distances, while compartmentalization within the cell itself might be another. His own work, using very similar tools as Reiter and Xu’s, has shown that in addition to a spatial encoding of information that distinguishes signals from the cilia or, for instance, from endosomes, cells can encode signaling information temporally. For instance, cAMP generated from the plasma membrane is short-lived, while endosomal cAMP is sustained.
There is much to learn about how organization within individual cells helps impart different signaling outcomes from a shared signal amplifier like cAMP. Also, while the conclusions about the HH pathway are at the individual cell level, it is in fact a multicellular response, as cells communicate with one another to eventually produce the slated tissue- and organism-level developmental changes. It is curious that an organelle such as the cilium can help cells talk to each other, and it suggests that cells can listen to multiple conversations simultaneously and integrate multiple circuits together to produce unique downstream signaling outcomes.
Researchers gathered to discuss trends and technologies that will enable biological research
Learn More
Three generations of scientists visit CZ Biohub SF to learn metagenomic sequencing techniques and analysis
Learn More
CZ Imaging Institute scientists mark milestone achievement with annotation of over 13,000 tomograms in just 3.5 days
Learn More
Stay up-to-date on the latest news, publications, competitions, and stories from CZ Biohub.
Cookies and JavaScript are required to access this form.