“Macrophage memory emerges from coordinated transcription factor and chromatin dynamics”
Wang, A.G.*; Son, M.*; Gorin, A.; Kenna, E.; Padhi, A.; Keisham, B.; Schauer, A.; Hoffmann, A.; Tay, S†
Cell Systems (accepted)
Microfluidic device for tissue culture and propagation of immune signaling in tissue cells
Microfluidic device for tissue culture and propagation of immune signaling in tissue cells
Cells within tissue respond to and secrete various biochemical signals to communicate local disease information and coordinate immune responses at both the tissue and organism levels. Although the spatial and temporal dynamics of signaling molecules contain valuable insights into disease progression and the efficacy of immune responses, these dynamics can vary significantly depending on the disease context, tissue state, stochastic cell responses, and from patient to patient. To overcome this complexity and enhance our understanding of the core mechanisms, we aim to collect multimodal data from tissues and deduce accurate — yet intuitive — models of pathogenesis or carcinogenesis under various conditions. To achieve our goal, we are developing advanced microfluidic platforms to quantify immune signaling and response dynamics in primary tissues (“microtissues”), and are integrating these empirical results with various mathematical approaches, including but not limited to network simulation, information theory, chaos theory, and deep learning. Ultimately, we will endeavor to identify a set of parameters most indicative of disease and tissue state, and formulate optimal therapeutic strategies for treating autoimmune diseases and cancer.
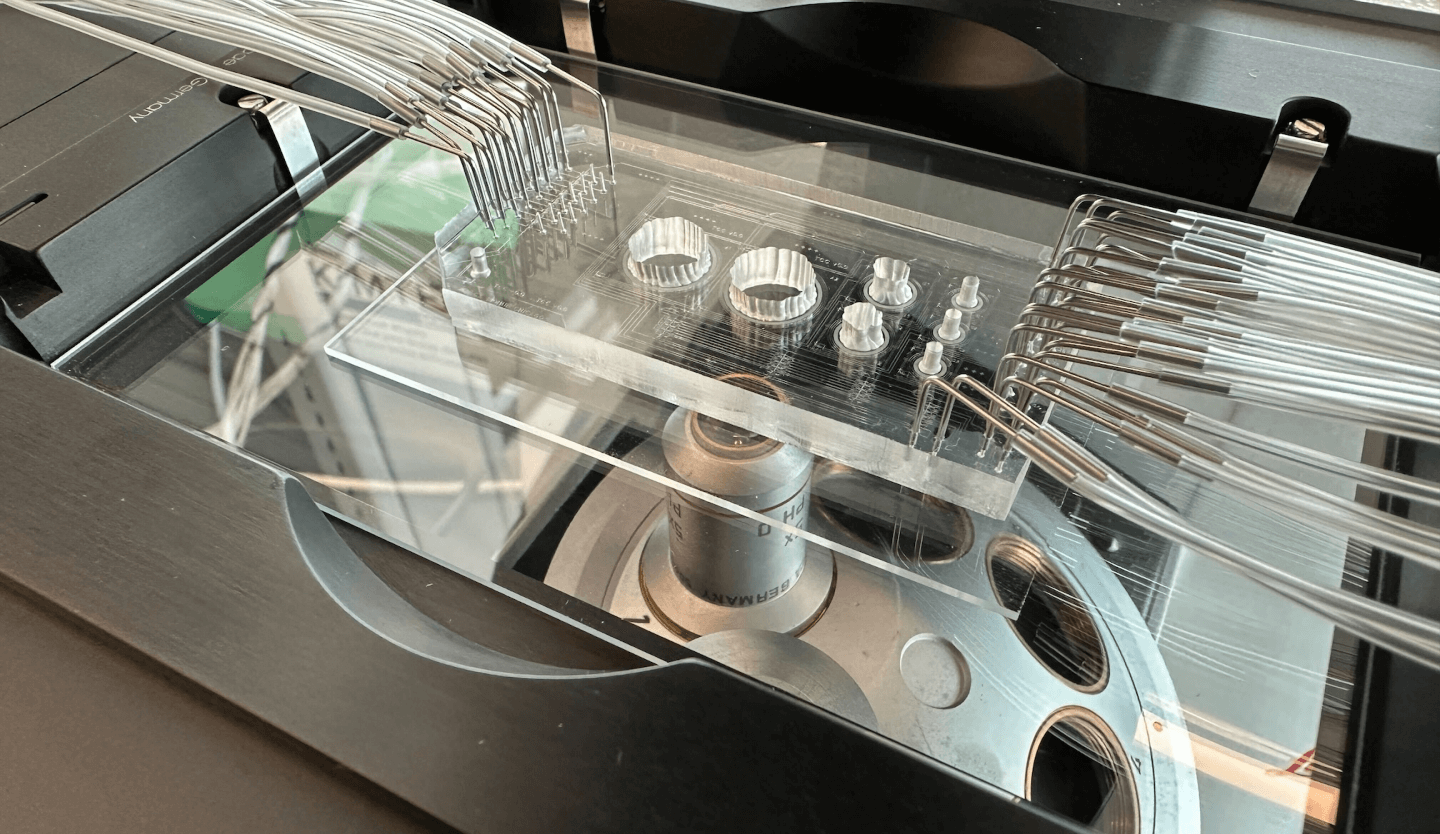
Microfluidic device for skin tissue and sequential secretome measurement
Virtual tissue: Simulating diffusion of pathogenic stimuli and immune signaling in 3-D tissue
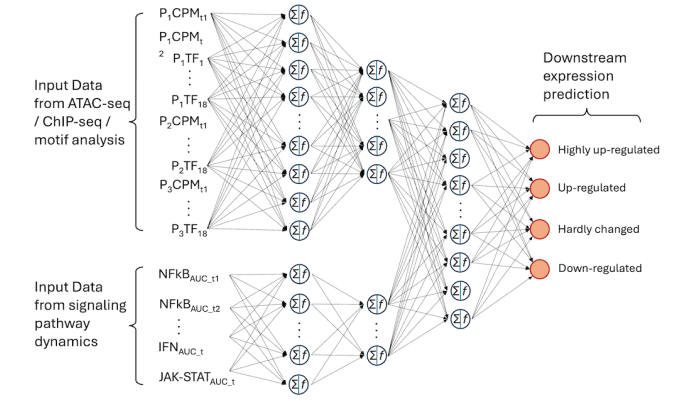
Neural network analysis for RNA-seq/proteomics prediction using multifaceted data
*: First author; †: Corresponding author; Underlined: Affiliated with CZ Biohub

Wang, A.G.*; Son, M.*; Gorin, A.; Kenna, E.; Padhi, A.; Keisham, B.; Schauer, A.; Hoffmann, A.; Tay, S†
Cell Systems (accepted)