At the intersection of machine learning and life science
At the intersection of machine learning and life science
In April 2021, a jail in a California County was experiencing a modest but stubborn COVID-19 outbreak, despite a rigorous screening protocol, a quarantine program, and other measures. The county health department wanted to know what was driving the spread – was it due mainly to new people coming into the jail or from ongoing transmission within the facility?
With help from scientists at CZ Biohub San Francisco – and a sophisticated cloud-based tool called CZ GEN EPI, jointly developed by CZ Biohub and the Chan Zuckerberg Initiative, that analyzes the genomic “fingerprints” of viruses – health officials were able to trace the transmission pathways, and they soon had an answer: their intake screening and quarantine protocols were effective; it was the infection control procedures that needed tightening.
“This was actually quite a surprise to the public health department, but it gave them their marching orders,” says Patrick Ayscue, an epidemiologist and senior biosecurity fellow at CZ Biohub SF. “Every health department is operating with limited resources and has to make trade-offs, so any time we’re able to help them target where they’re putting their resources is a win.”
Genomic epidemiology is a powerful scientific tool that the Centers for Disease Control and Prevention (CDC) and a few large state agencies have in their arsenal to trace outbreaks of infections such as tuberculosis and E. coli or other food-borne pathogens. But traditionally it has been out of reach for most local health jurisdictions and public health practitioners. So in 2020, as it became apparent that COVID-19 was becoming a full-blown pandemic, Ayscue and scientists at CZ Biohub stepped in – first to provide the service as a stopgap, and then to empower dozens of state and local health agencies in California and elsewhere to do genomic epidemiology themselves – to sequence and analyze SARS-CoV-2-positive samples in order to generate actionable insights.
“We ended up working with 22 county health departments in California and generating more than 13,000 SARS-CoV-2 genome sequences, supported hundreds of outbreak investigations, resulting in probably dozens of policy changes across the groups we were working with,” he says.

CZ Biohub SF scientist Patrick Ayscue gave a training in genomic epidemiology last month in Bangkok, Thailand.
Now CZ Biohub SF is going international with its training. In collaboration with the Association of Public Health Laboratories and the CDC, Biohub scientists traveled to Bangkok last month to train lab personnel from throughout Southeast Asia and help them establish genomic epidemiology programs in their own laboratories. They’re interested in tracing antimicrobial-resistant pathogens (sometimes called “superbugs”), dengue fever, Japanese encephalitis, respiratory pathogens, hepatitis, and tuberculosis. “We expect they will be able to start generating their own analyses to inform outbreak response and policy formation in their home countries,” Ayscue says.
And in the U.S. many of the local public health practitioners have gone on to leverage their training to develop full-fledged programs of their own, enabling their officials to better understand and respond to public health issues in their own jurisdictions, whether it’s sexually transmitted infections, hospital-acquired infections, food-borne pathogens, or localized diseases, such as Valley fever, which is a serious concern in parts of California.
“Before we were able to do our own sequencing, we were essentially at the mercy of other laboratories. Whether or not a specific patient or population or sample type was a priority for them dictated whether or not it would ultimately even get sequenced, and it would take weeks and weeks to receive results,” says Denise Lopez, the public health lab director for Tulare County in California’s Central Valley, which has one of the first labs to receive training from CZ Biohub SF experts.
“By doing our own genomic sequencing we can control what gets sequenced, what populations get priority, how intensively we want to sequence, and what percentage of positives we want to look at,” says Lopez. “With COVID-19 we did runs once a week, so essentially all of our sequencing was up to date within 7 to 14 days, which was phenomenal and really not seen anywhere else in the state.”
Genomic epidemiology allows practitioners to infer patterns of a disease outbreak by analyzing the genetic sequence data of pathogens. It relies on the fundamental principle that pathogens evolve at a timescale similar to the timescale at which infectious disease spreads through populations.
When pathogens spread from person to person, there will be errors as they replicate. These genetic mutations may be beneficial, neutral, or harmful for the pathogens, but they make it possible to plot patterns of shared ancestry and create a phylogenetic tree. That information allows public health officials to then answer questions such as what viral variants are present, which cases share a common transmission chain, where transmission is occurring, and whether interventions are working effectively.
But being able to do genomic epidemiology normally requires both specialized training and sophisticated equipment and infrastructure – sequencing machines, reagents, data management infrastructure, and analytical skills, to name a few.
CZ GEN EPI, a free, open-source genomic epidemiology analysis platform, reduces the analytical skills needed by users. It allows users to upload genomic data and automatically builds phylogenetic trees to trace variants and outbreaks with no coding required.
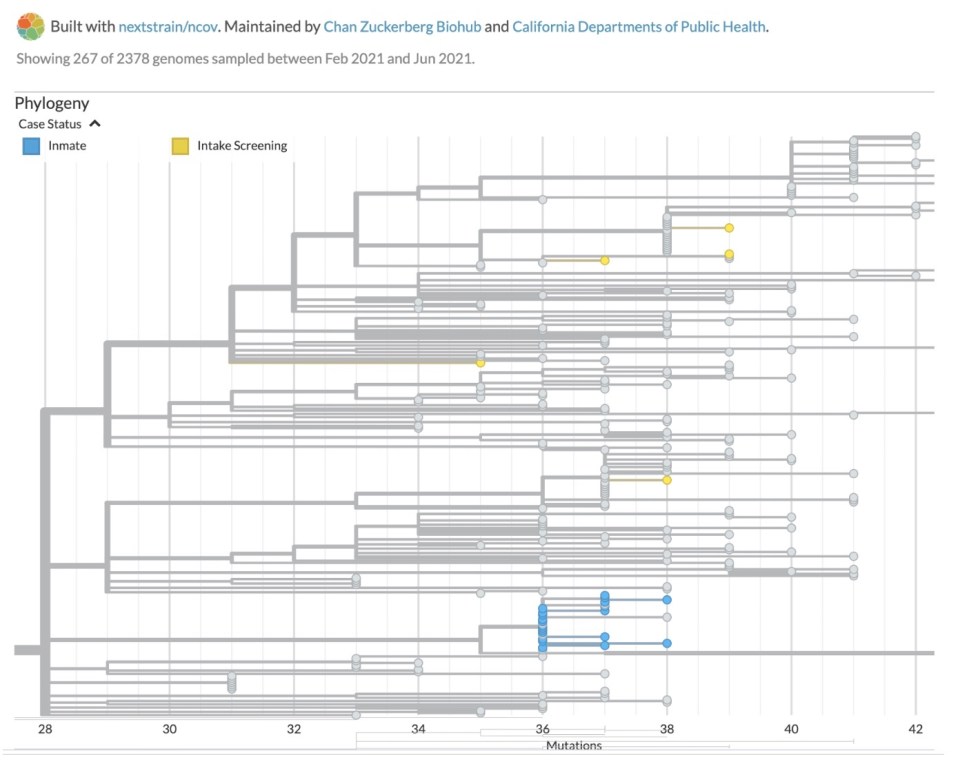
A phylogenetic tree helps show how viruses spread and evolve over time.
“Putting these capabilities in the hands of states and local health departments, where the response is actually happening, unlocks a lot of functionality and capabilities that they wouldn’t otherwise have, particularly for infections that are happening on the order of months-long timescales,” Ayscue says.
Lopez says genomic epidemiology “was not on our radar at all” when COVID hit. “Biohub walked us through the entire thing, and we were able to get up and running within just a matter of weeks,” she says.
Being able to do their own sequencing of SARS-CoV-2 was immensely helpful, Lopez says. For example, they were able to understand the impacts of various mutations and learned that whenever a particularly concerning variant would show up in bigger cities, it would arrive in their county within two to three weeks.
“It helped us to be able to plan. We were able to estimate how long it likely would take before it hit our area, which is very helpful for a variety of reasons,” she says. “It told us we should ramp up the number of samples we were sequencing in order to improve our sensitivity of detecting those variants, and also it helped our hospitals to anticipate that there could be some additional waves coming.”
Almost all of the groups that CZ Biohub SF trained are continuing to use genomic epidemiology, whether with CZ GEN EPI or another platform, and are starting to look at pathogens besides SARS-CoV-2.
“Pretty much across the board they’ve been able to see the value that genomic epidemiology is bringing to their work,” says Ayscue. “I don’t see anybody planning on walking away from it or not sustaining it.”
Another jurisdiction that scientists from CZ Biohub and CZI worked with was Humboldt County, a rural area in northern California. They helped public health officials there to trace an outbreak linked to a farm and another one in a skilled nursing facility. Like the jail example, those cases demonstrated the ability of genomic epidemiology to inform response; the Humboldt cases were published in the journal BMC Public Health last year.
Tulare County has now moved onto another genomic epidemiology platform set up by the state of California, but Lopez credits CZ Biohub SF with showing the value of genomic epidemiology to her county. “Generally genomic sequencing is not a revenue-generating test. You’re usually not billing insurance for it, so it really requires a specific grant to fund it or buy-in from leadership.”
Lopez has many plans for how they’ll use their genomic sequencing capabilities now that the COVID-19 pandemic is winding down, including for food-borne illness surveillance, antimicrobial resistance monitoring, and more comprehensive surveillance of respiratory illnesses.
“We learned we didn’t have much baseline data on what viruses are circulating in a ‘normal’ year, and what would be considered a high rate of any particular respiratory illness. We’re hoping to dig deeper into that, so it can inform our preparedness and allow us to anticipate spikes early,” she says. “In my lab genomic sequencing is now viewed as a good investment that can drive public health decisions.”
At the intersection of machine learning and life science
Learn More
Unraveling the mystery of brain infections
Learn More
Piecing together new technologies for biomedicine
Learn More
Stay up-to-date on the latest news, publications, competitions, and stories from CZ Biohub.
Marketing cookies are required to access this form.