Obsessed with the nucleus
Obsessed with the nucleus
Countless diseases are caused by genetic mutations that lead to dysfunctional proteins, and even minor disruptions in the folding of a protein can affect its ultimate structure and shake up the cell’s delicate balancing act. In a well-known case, a single mutation in a 1,480-amino-acid ion channel protein called CFTR can lead to cystic fibrosis, while mutations in certain other proteins can cause them to form alternative structures that have been implicated in several neurodegenerative conditions including Parkinson’s and Alzheimer’s diseases.
And as the changes in the Spike protein in Delta and other SARS-CoV-2 variants have powerfully demonstrated, it is important to understand how mutations affect protein function in infectious disease agents.
To gain a quantitative understanding of how such mutations can affect all aspects of a protein’s function, Chan Zuckerberg Biohub Investigator Polly Fordyce, jointly with Dan Herschlag’s group and other colleagues at Stanford University, has developed a platform called HT-MEK (High-Throughput Microfluidic Enzyme Kinetics), which can quickly and precisely monitor the effects of more than 1,500 point mutations on a number of key activity parameters of a protein of interest.
In work led by Craig Markin, a postdoc in the Fordyce and Herschlag labs, when applied as a proof of principle to an enzyme known as PafA (structure pictured above), the HT-MEK device allowed the team to identify regions of the protein that are sensitive to mutation and which functions are affected. As reported in the July 23, 2021 issue of Science, in addition to exploring mutational effects on PafA’s active site—the business end of enzymes, directly involved in binding the substrate upon which they act and the chemical transformations they catalyze—the technique also revealed a surprisingly large number of mutations far removed from the active site, including so-called allosteric sites, that disturb function.
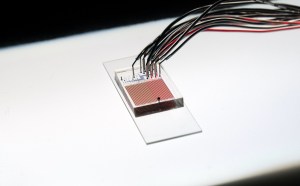
The HT-MEK device is small enough to fit on a 1″ x 3″ slide. (Courtesy of P. Fordyce)
Combined with assessments of how faithfully the mutated enzymes are folded, the work also allowed the team to tease apart the effects mutations have on function from the effects they have on misfolding and unfolding.
“We already have a lot of cellular assays to assess the consequences of different mutations in a cell, but these results can be really convolved,” says Fordyce. “If you see a protein with 10% activity, it could mean that 10% of the protein is folded but 100% of what’s folded is active. Or it could be that it’s 100% folded but it has only 10% of wild-type activity.”
What has been lacking, she believes, is a way to link structure and function. “Teams like Deep Mind have developed tools like AlphaFold 2 that are incredible at predicting the low-energy structure of a protein from its amino acid sequence, but we can’t yet predict how that sequence encodes function,” she says. Indeed, enzymes can adopt essentially the same fold, but perform very different chemistries or do so with varying levels of success, so “the structure itself doesn’t encode all of the information that we need in order to solve that next problem.”
In the crowded environment of a cell, mutations that disrupt any of the many crucial exchanges that take place—be they protein–protein interactions, chemical reactions, or movement through organelles—could be disastrous.
Among the most important proteins in the cell’s house of cards are enzymes, which carry out the chemical reactions that make life possible. Where each enzyme forms a tiny room of its own in the house of the cell, its amino acid sequence, three-dimensional structure, and conformational dynamics are the framework.
Fordyce, Herschlag, and colleagues used the HT-MEK platform to monitor the activity of more than 1,000 mutants of PafA, a highly stable prototype bacterial alkaline phosphatase whose mechanism is well-characterized, to explore how mutations within and beyond the active site affect function. They chose PafA because of its large dynamic range for facilitating its chemical reaction, the better with which to measure even the smallest changes in the variants.
Highlighting the ever-shrinking technology of collecting data at scale, HT-MEK consists of a small chip with microfluidics at its core. It’s the size of about two 25-cent coins and has an elaborate and mostly automated system of chambers and valves. Across one chip, more than 670,000 enzymatic reactions could be performed on GFP-tagged versions of individual PafA variants immobilized in individual chambers.
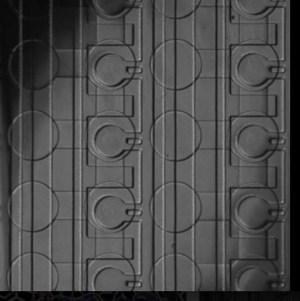
The device is a network of channels formed in two slabs of a polymer stacked on top of each other. Pneumatic valves form where channels cross each other in different layers: pressurizing the top channel deflects a membrane to restrict flow in the bottom channel (just like stepping on a garden hose), an application of the “Quake valve” technology originally developed in the Stanford lab of CZ Biohub Co-President Steve Quake. (Courtesy of P. Fordyce)
The authors made at least two individual mutations at each of the protein’s 526 amino acid residues, and for each one they measured a battery of kinetic and thermodynamic parameters that report in parallel on reaction speeds and energetics across multiple substrates and inhibitors, and they could dissect out folding effects from catalytic effects.
“The power of this approach is that you only make a protein once and then you can probe it under all these conditions allows you to get all of the phenotypic information you need to quantitatively describe the enzyme,” says Fordyce.
Over 70% of the PafA mutations, including many far outside the active site, diminished the activity of the enzyme. The research team could explain nearly a third of those by showing that they adopted a long-lived misfolded form that is still kinetically stable. “If we were doing a deep-scanning mutagenesis experiment or another traditional experiment where you couldn’t tease apart activity and folding, we wouldn’t have been able to say that that’s what a lot of these mutations are doing,” Fordyce says.
Many of the variants that affect the enzyme’s activity cluster around the active site, and a map of the relative impacts of changing each residue revealed stretches of the protein where mutations affect individual reaction steps, including regions that affect hydrolysis rates against two different types of substrate.
Nonetheless, the team’s results lend further support to the view that enzymatic function goes beyond the active site, such that perturbing the structure at distant allosteric sites—either with mutations or with small-molecule binders—can affect the active site structure to affect overall activity.
Results from mutations in one stretch of amino acids to change the charges in the so-called “nucelophile helix” (seen in green in the illustration of PafA’s structure above), point to an evolutionary history for PafA that shaped it to be such an efficient enzyme, where it’s necessary to destabilize its lowest energy state compared to the higher catalytic “transition” state by repelling the charged reaction product, Pi. This support network of residues act like a boxer’s cornerman, stabilizing the helix to maintain the charged repulsion.
When applied to disease-linked enzymes, these insights could allow us to classify rare variants that cause disease into subtypes to better guide therapy, to design new therapeutics at more specific sites beyond the active sites, and to make designer enzymes that have unique functions.
For Fordyce, devices like HT-MEK signal an era of more quantitative descriptions of biology that she likens to the advances in weather prediction made possible when atmospheric measurements could be obtained in absolute, rather than relative, terms—by measuring temperature, humidity, and other absolute physical constants.
“In terms of thinking about enzymes, we have little bits of those data from traditional site-directed mutagenesis. Deep-scanning mutagenesis just gives us relative changes,” says Fordyce. “It’s not getting information in the language of the physical constants, and that’s what HT-MEK allows us to do, at scale.”
Obsessed with the nucleus
Learn More
A special visit with a nonagenarian expert in pathogenic amoebae
Learn More
Unraveling the mystery of brain infections
Learn More
Stay up-to-date on the latest news, publications, competitions, and stories from CZ Biohub.
Marketing cookies are required to access this form.