Leonetti Group: Intracellular Architecture
How are cells internally organized in space and time? We are mapping the wiring diagram of the human proteome to unravel the changes at play when diseases strike.
Our multidisciplinary group of biologists, engineers, and data scientists strives to understand how human cells are built. We seek to characterize fundamental mechanisms in physiology and disease, but also to “reverse engineer” the cell—to understand the details of how it is built in order to be able to predictably tune its properties and behavior. In particular, we develop and deploy technologies to illuminate the function of proteins within cells and manipulate them. One of our goals is to build open datasets, software, and protocols for the entire scientific community to use and explore.
-
 High-throughput cell engineering
High-throughput cell engineering- We develop CRISPR methods to generate genome-scale libraries of engineered human cell lines
- We build software and automation to streamline the design, execution, and analysis of our experiments
-
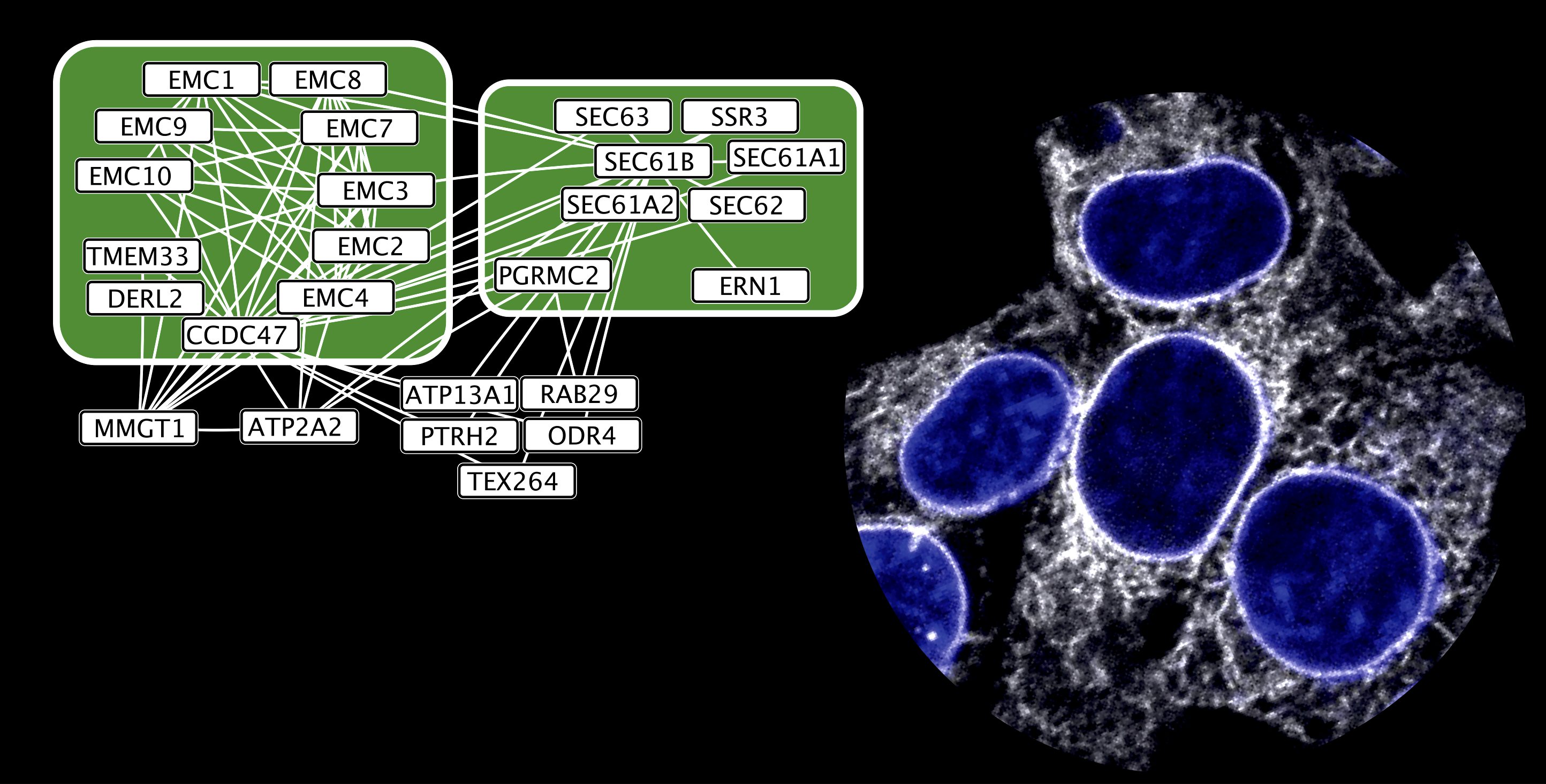 Multi-omics protein profiling
Multi-omics protein profiling- We use high-throughput live-cell fluorescence microscopy to map protein localization in space and time
- We apply proteomics mass spectrometry to profile protein–protein interactions and delineate functional networks
-
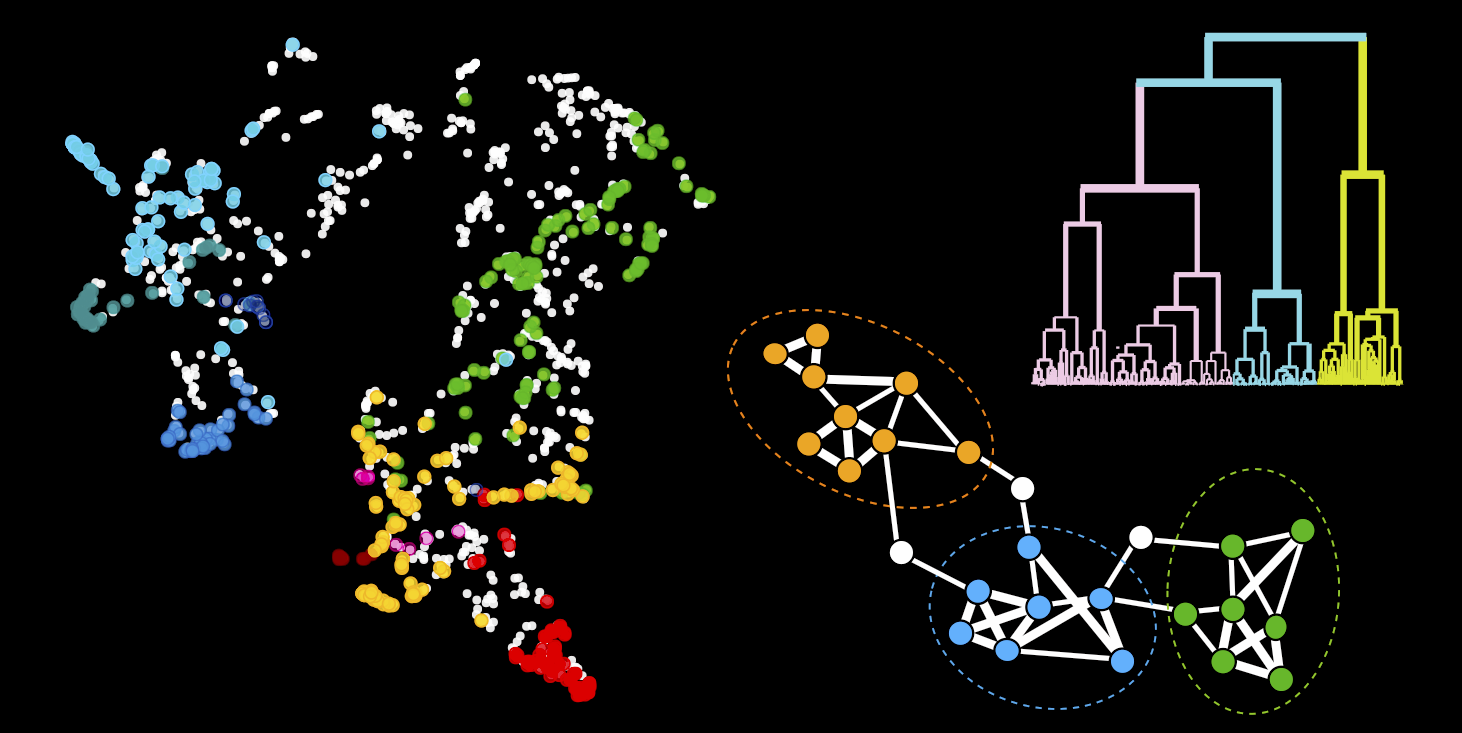 Collaborative data science
Collaborative data science- We develop self-supervised machine learning approaches for image analysis
- We build websites and open-source our code so that everyone can explore and reuse our data and analyses
 OpenCell—our flagship project—is a proteome-scale library of fluorescently engineered cell lines to define the localization and interactions of human proteins. We have data for over 1,300 proteins so far and we built a fully interactive website to share our data.
OpenCell—our flagship project—is a proteome-scale library of fluorescently engineered cell lines to define the localization and interactions of human proteins. We have data for over 1,300 proteins so far and we built a fully interactive website to share our data.
Start exploring at opencell.czbiohub.org.
More on our work in this recent presentation.


