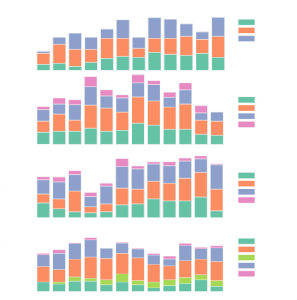
Illumina Technology
Library Structure
- Illumina Adapter Sequences: a comprehensive list of all Illumina adapters and indices sequences from various products (including nextera, truseq, etc.)
- Standard Index oligos:
- P7: 5’ CAAGCAGAAGACGGCATACGAGAT[index][adapter_sequence]3’
- P5: 5′ AATGATACGGCGACCACCGA[dual_index][adapter_sequence] 3′
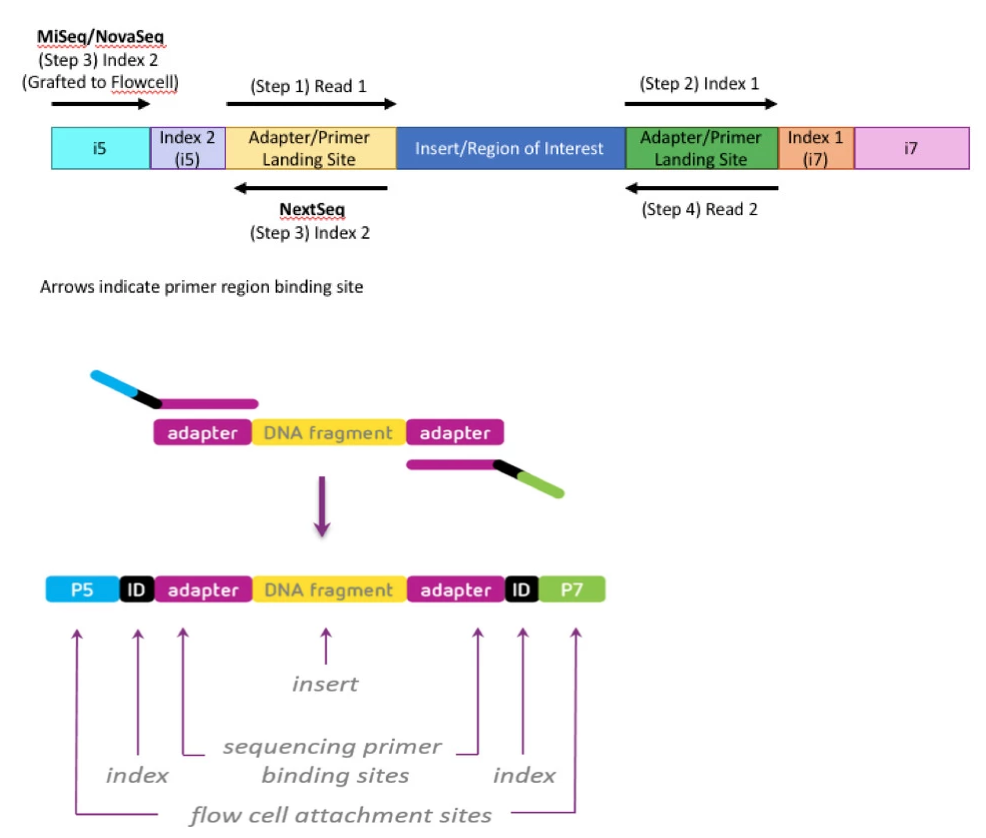
Laser and Dyes
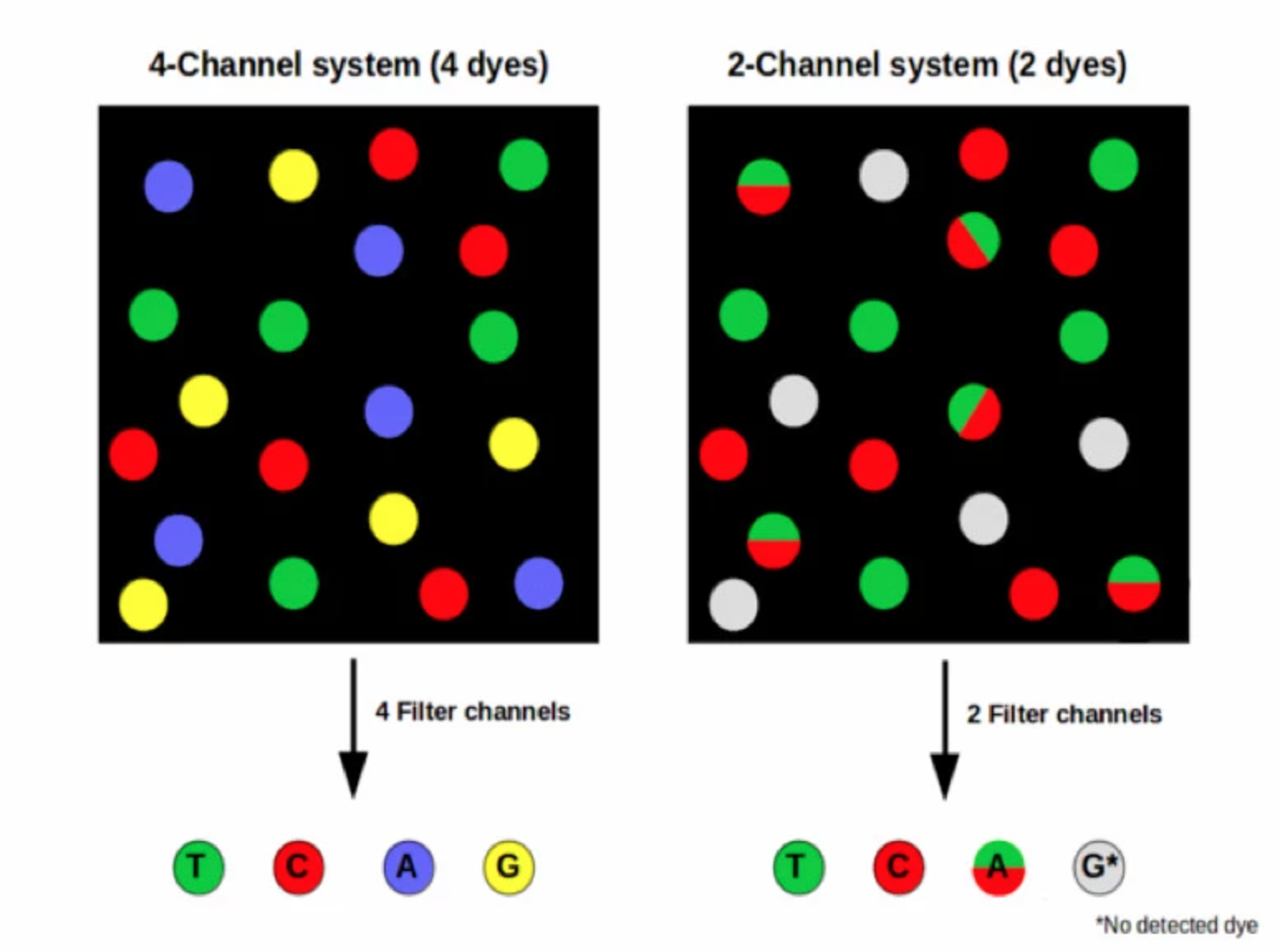
Nextseq and Novaseq – 2 dye
- Rather than a separate dye for each base, 2-channel SBS uses a mix of dyes. Images are taken of each DNA cluster using red and green wavelength filter bands.
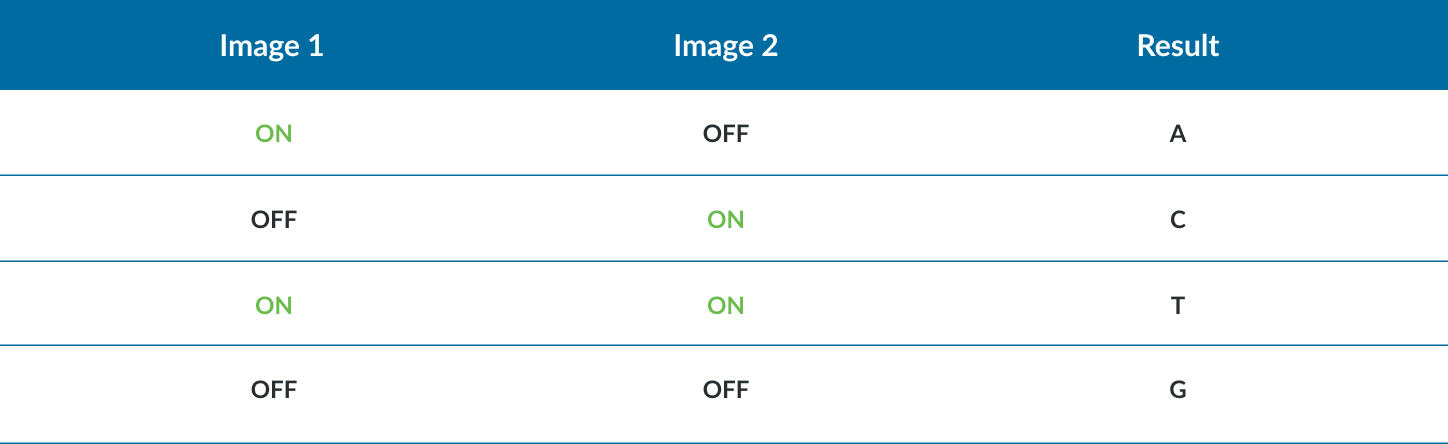
- Clusters seen in red or green images are interpreted as C and T bases, respectively. Clusters observed in both red and green images are flagged as A bases (appearing as yellow clusters), while unlabeled clusters are identified as G bases).
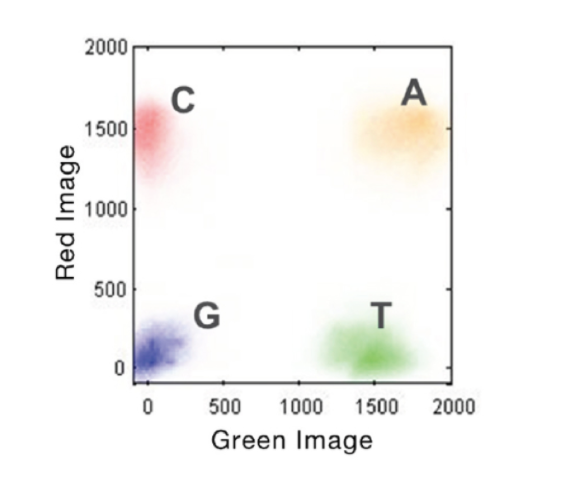
- Miseq – 4 dyes/2 lasers
- G/T share one laser and C/A share another. When choosing indexes, make sure each base has diversity between the two lasers.
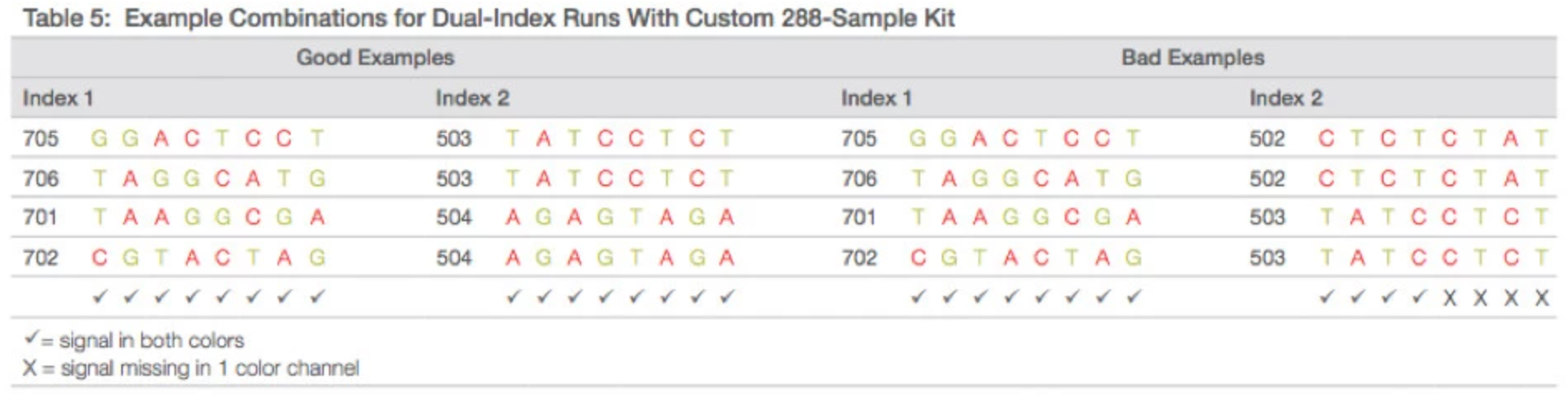
- iSeq
- One dye and two chemistry step
- For all sequencers: there is no index primer site for i7 in PhiX therefore spiking in more phix would not help with index diversity. Multiplexing under six samples requires careful selection of barcodes.
Illumina Sequencing Outputs
- For all sequencers, the minimum R1 cycle is 25 (26 for NovaSeq)
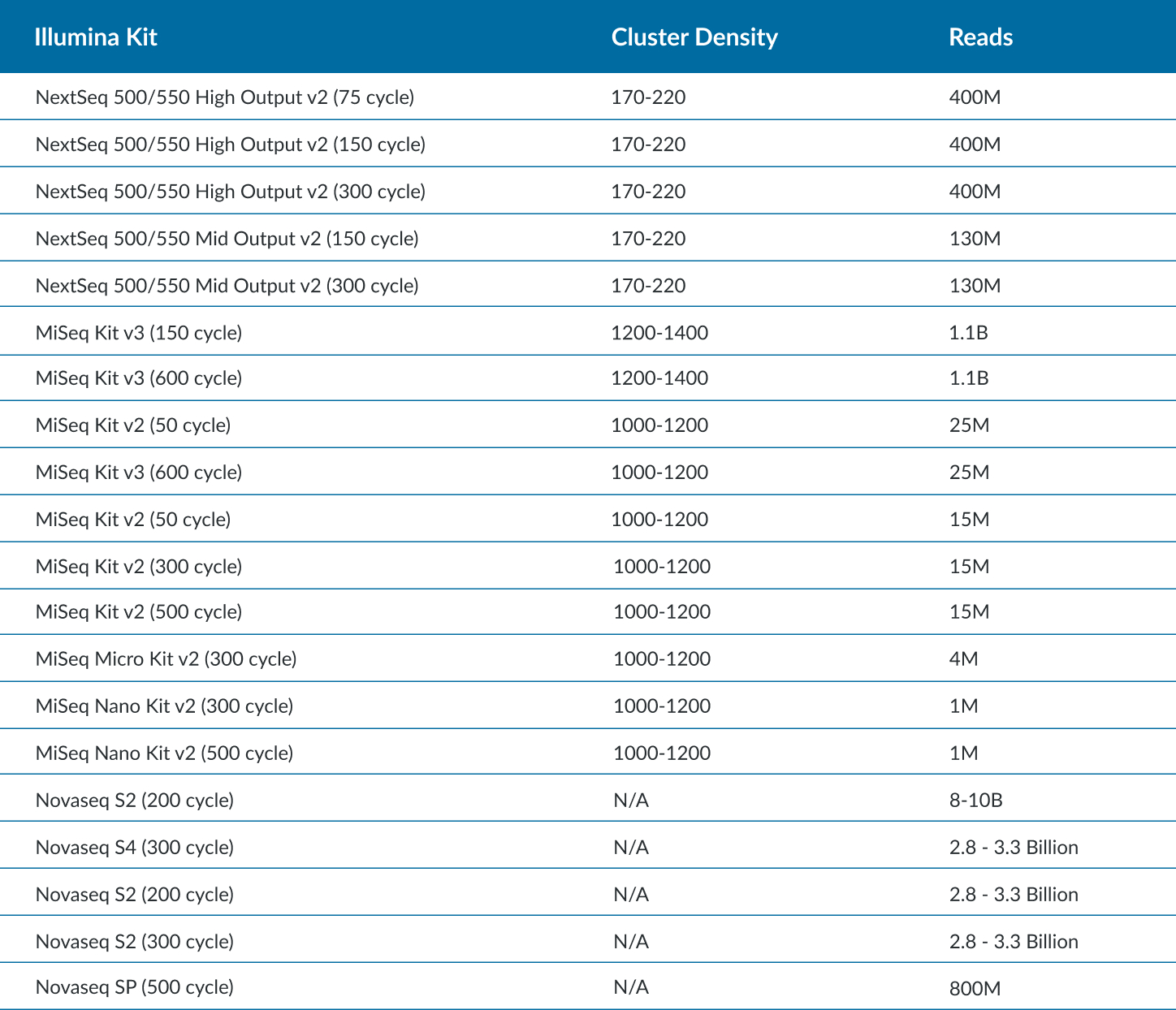
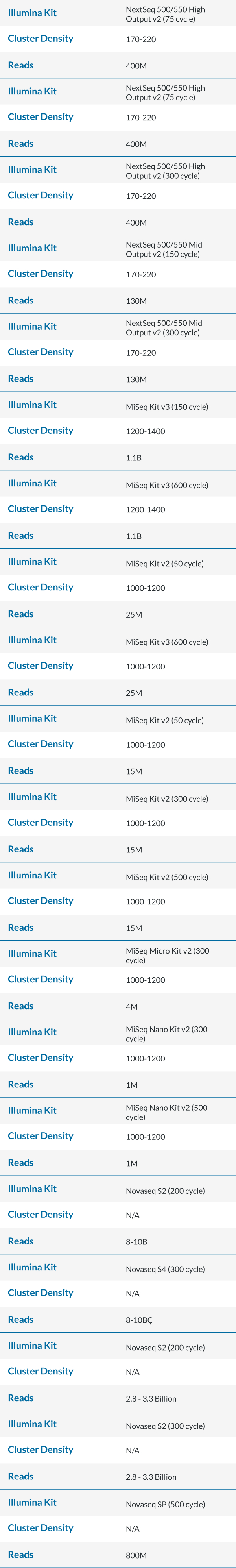
Maximum Cycles in a SBS Kit
- There is overage for extra cycles built into every kit. Operators could use each to the fullest extent by calculating the maximum cycles for read 1 and read 2. Please adjust accordingly, as some chemistry need additional cycles for dual index read (also known as dark cycling).
- Example: A NovaSeq S2 300 cycle kit has 325 cycles, 325 – 8 (index1) – 8 (index2) – 7 (dark cycles) = 302 cycles remaining for read 1 and read 2. This kit is validated for up to a paired end 151 cycles.
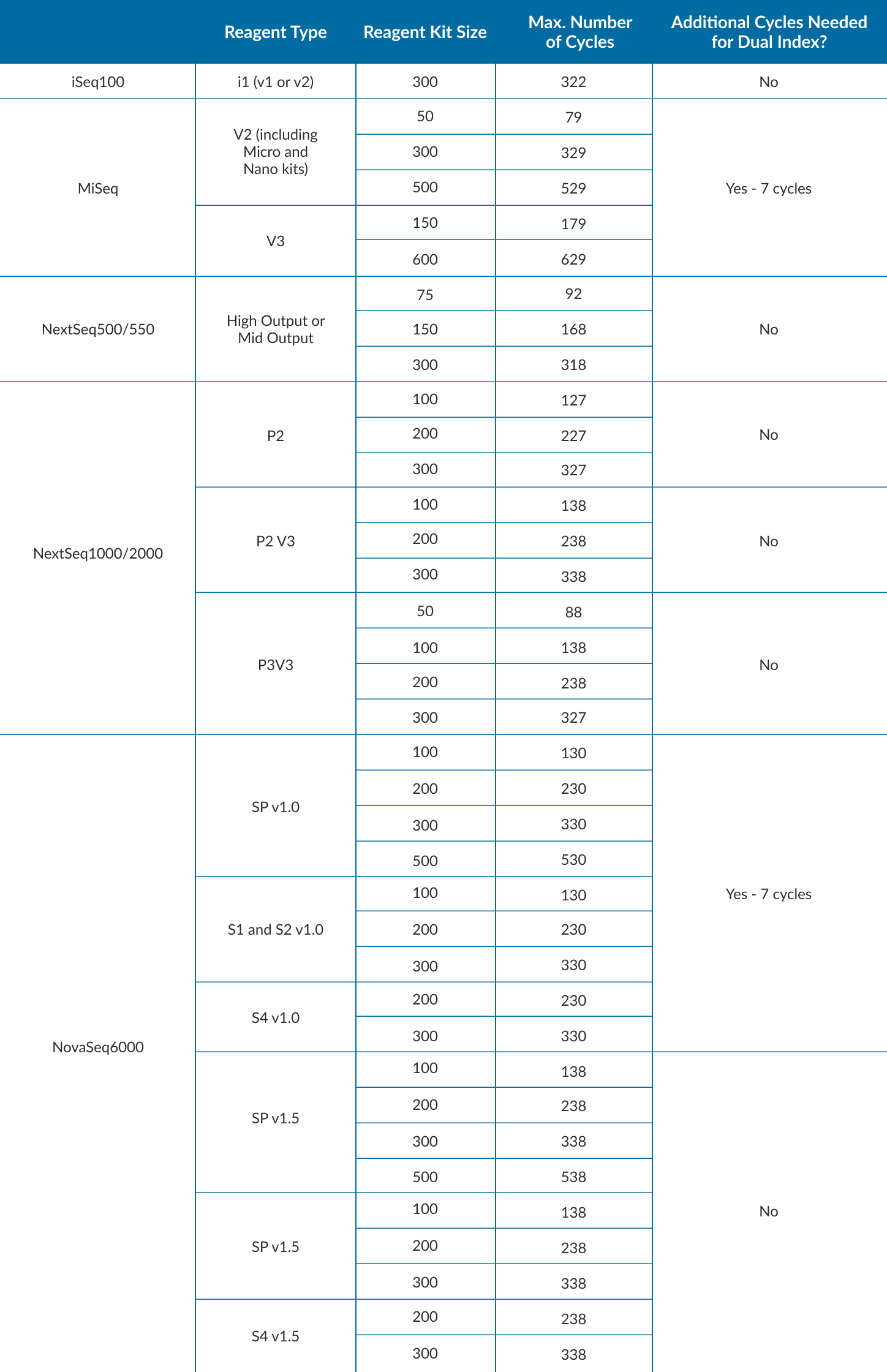

How Many Reads Do I Need?
- Depends on many things: input material, library prep, what biological question to answer.
- Start with the Illumina Sequencing Coverage Calculator to approximate coverage