Pete Farley
Director of Communications
The Chan Zuckerberg Biohub (CZ Biohub), a nonprofit medical research organization, today announced that it is awarding $13.7 million over three years to support cutting-edge biomedical research from seven teams of scientists, physicians, and engineers, with each team including faculty members from the University of California, San Francisco (UCSF); the University of California, Berkeley (UC Berkeley); and Stanford University.
The awards will fund two new programs: the CZ Biohub Microbiome Initiative and the CZ Biohub Intercampus Research Awards.
“For the first time, these new awards bring together highly talented investigators from all three campuses to collaborate on promising new approaches to major biomedical problems,” said Joe DeRisi, co-president of CZ Biohub. “By drawing on the strengths of all three institutions, we believe these teams will accomplish what is now beyond the reach of individual investigators.”
CZ Biohub Microbiome Initiative
Launched as a pilot program earlier this year, the CZ Biohub Microbiome Initiative provides $4 million over three years to carry out research on the community of microbes within the human body that influence many aspects of health, from nutrition and immune function to drug metabolism. The Microbiome Initiative brings together eight leading microbiome experts from all three campuses based on their highly complementary and synergistic research interests.
CZ Biohub Intercampus Research Awards
Assembling the Microbiome Initiative team inspired CZ Biohub to create the Intercampus Research Awards. The new competitive awards program promotes collaborative research by bringing together clinicians, biologists, chemists, data scientists, mathematicians, engineers, and bioethicists in teams that each include faculty members from all three campuses. The competition drew applications from 83 teams. CZ Biohub initially planned to support three teams but was inspired to increase the number of awards to six, providing $9.7 million over three years.
“This new collaborative team-based funding allows investigators across the three campuses to tackle demanding problems to enhance health,” said Steve Quake, co-president of CZ Biohub. “These research teams will shed new light on a diverse and challenging set of questions that will advance our understanding while developing technologies that open fresh avenues of research.”
Statement from Priscilla Chan & Mark Zuckerberg, Co-Founders, The Chan Zuckerberg Initiative
“We launched the Biohub to bring together some of the brightest scientific minds in the Bay Area with world-class engineering teams, in order to help accelerate the pace of discovery and make faster progress in the fight against disease. Just two years after its launch, it is incredible to see how the Biohub has helped spark promising new collaborations, tools, and research to enable and empower the entire scientific community. Breakthroughs occur when researchers work together, within and across disciplines. By creating even more opportunities to do so through these two new programs, we believe we’ll get closer to our goal of curing, preventing, or managing all disease by the end of the century,” said Priscilla Chan and Mark Zuckerberg, co-founders of The Chan Zuckerberg Initiative.
Statements from Stanford, UC Berkeley, and UCSF
UC Berkeley Chancellor Carol Christ said, “We are thrilled by the extent to which these awards honor and support the notion that our most pressing challenges can only be surmounted by transcending the lines that have too long divided academic disciplines, departments, and even institutions. The awards will also help support our efforts to extend the reach of world-class, fundamental research through initiatives that can speed the translation of discoveries into inventions and services for the benefit of all.”
UCSF Chancellor Sam Hawgood said, “With its unique model of multi-institutional collaboration, the Chan Zuckerberg Biohub has become a keystone of the Bay Area ecosystem of innovation. The new research awards program and the Microbiome Initiative further strengthen the scientific and technological bonds Biohub has already forged among UCSF, Stanford, and UC Berkeley. We look forward to working with our partner institutions on these exciting new projects to advance knowledge and human health.”
Stanford University President Marc Tessier-Lavigne said, “I am excited that the Chan Zuckerberg Biohub has chosen to fund innovative, high-risk research with great potential for enhancing our understanding of human health. By supporting teams that are collaborating across disciplines and across our three institutions, the Biohub will enable scientific breakthroughs that increasingly require insights from multiple disciplines and perspectives.”
APPENDIX 1
|
Investigator |
Institution |
Program |
Team |
Role |
|
Christopher Lowe |
Stanford |
Intercampus Research Awards |
Beyond model systems: insights into genome evolution and cellular innovations |
Project Leader |
|
Wallace Marshall |
UCSF |
Intercampus Research Awards |
Beyond model systems: insights into genome evolution and cellular innovations |
Project Leader |
|
Stephen Palumbi |
Stanford |
Intercampus Research Awards |
Beyond model systems: insights into genome evolution and cellular innovations |
Project Leader |
|
Daniel Rokhsar |
Berkeley |
Intercampus Research Awards |
Beyond model systems: insights into genome evolution and cellular innovations |
Project Leader |
|
Irving Weissman |
Stanford |
Intercampus Research Awards |
Beyond model systems: insights into genome evolution and cellular innovations |
Project Leader |
|
Ayelet Voskoboynik |
Stanford |
Intercampus Research Awards |
Beyond model systems: insights into genome evolution and cellular innovations |
Collaborator |
|
Laurent Coscoy |
Berkeley |
Intercampus Research Awards |
Defining host responses of virus-infected and uninfected neighbor cells |
Project Leader |
|
Karla Kirkegaard |
Stanford |
Intercampus Research Awards |
Defining host responses of virus-infected and uninfected neighbor cells |
Project Leader |
|
Melanie Ott |
UCSF Gladstone Institutes |
Intercampus Research Awards |
Defining host responses of virus-infected and uninfected neighbor cells |
Project Leader |
|
Peter Sarnow |
Stanford |
Intercampus Research Awards |
Defining host responses of virus-infected and uninfected neighbor cells |
Project Leader |
|
Nir Yosef |
Berkeley |
Intercampus Research Awards |
Defining host responses of virus-infected and uninfected neighbor cells |
Collaborator |
|
John C. Boothroyd |
Stanford |
Intercampus Research Awards |
Imaging complex biological machines in action |
Project Leader |
|
Wah Chiu |
Stanford |
Intercampus Research Awards |
Imaging complex biological machines in action |
Project Leader |
|
Carolyn Larabell |
UCSF |
Intercampus Research Awards |
Imaging complex biological machines in action |
Project Leader |
|
James A. Sethian |
Berkeley |
Intercampus Research Awards |
Imaging complex biological machines in action |
Project Leader |
|
Mark Le Gros |
UCSF |
Intercampus Research Awards |
Imaging complex biological machines in action |
Collaborator |
|
W.E. Moerner |
Stanford |
Intercampus Research Awards |
Imaging complex biological machines in action |
Collaborator |
|
Russ B. Altman |
Stanford |
Intercampus Research Awards |
Machine learning for interpreting rare genetic variation in comprehensive newborn screening and pharmacogenetics |
Project Leader |
|
Steven E. Brenner |
Berkeley |
Intercampus Research Awards |
Machine learning for interpreting rare genetic variation in comprehensive newborn screening and pharmacogenetics |
Project Leader |
|
Carlos Bustamante |
Stanford |
Intercampus Research Awards |
Machine learning for interpreting rare genetic variation in comprehensive newborn screening and pharmacogenetics |
Project Leader |
|
Renata C. Gallagher |
UCSF |
Intercampus Research Awards |
Machine learning for interpreting rare genetic variation in comprehensive newborn screening and pharmacogenetics |
Project Leader |
|
Kathleen M. Giacomini |
UCSF |
Intercampus Research Awards |
Machine learning for interpreting rare genetic variation in comprehensive newborn screening and pharmacogenetics |
Project Leader |
|
Michael I. Jordan |
Berkeley |
Intercampus Research Awards |
Machine learning for interpreting rare genetic variation in comprehensive newborn screening and pharmacogenetics |
Project Leader |
|
Jodi Halpern |
Berkeley |
Intercampus Research Awards |
Machine learning for interpreting rare genetic variation in comprehensive newborn screening and pharmacogenetics |
Collaborator |
|
Barbara Koenig |
UCSF |
Intercampus Research Awards |
Machine learning for interpreting rare genetic variation in comprehensive newborn screening and pharmacogenetics |
Collaborator |
|
David Magnus |
Stanford |
Intercampus Research Awards |
Machine learning for interpreting rare genetic variation in comprehensive newborn screening and pharmacogenetics |
Collaborator |
|
Jennifer Puck |
UCSF |
Intercampus Research Awards |
Machine learning for interpreting rare genetic variation in comprehensive newborn screening and pharmacogenetics |
Collaborator |
|
Jillian Banfield |
Berkeley |
Microbiome Initiative |
Microbiome Initiative |
CZBMI Investigator |
|
Michael Fischbach |
Stanford |
Microbiome Initiative |
Microbiome Initiative |
CZBMI Investigator |
|
Kerwyn (KC) Huang |
Stanford |
Microbiome Initiative |
Microbiome Initiative |
CZBMI Investigator |
|
Alexander (Sandy) Johnson |
UCSF |
Microbiome Initiative |
Microbiome Initiative |
CZBMI Investigator |
|
Katherine Pollard |
UCSF Gladstone Institutes |
Microbiome Initiative |
Microbiome Initiative |
CZBMI Investigator |
|
David Relman |
Stanford |
Microbiome Initiative |
Microbiome Initiative |
CZBMI Investigator |
|
Justin Sonnenburg |
Stanford |
Microbiome Initiative |
Microbiome Initiative |
CZBMI Investigator |
|
Peter Turnbaugh |
UCSF |
Microbiome Initiative |
Microbiome Initiative |
CZBMI Investigator |
|
Rima Arnaout |
UCSF |
Intercampus Research Awards |
Multi-scale deep learning and single-cell models of cardiovascular health |
Project Leader |
|
Euan Ashley |
Stanford |
Intercampus Research Awards |
Multi-scale deep learning and single-cell models of cardiovascular health |
Project Leader |
|
J. Ben Brown |
Berkeley |
Intercampus Research Awards |
Multi-scale deep learning and single-cell models of cardiovascular health |
Project Leader |
|
Atul Butte |
UCSF |
Intercampus Research Awards |
Multi-scale deep learning and single-cell models of cardiovascular health |
Project Leader |
|
James Priest |
Stanford |
Intercampus Research Awards |
Multi-scale deep learning and single-cell models of cardiovascular health |
Project Leader |
|
Bin Yu |
Berkeley |
Intercampus Research Awards |
Multi-scale deep learning and single-cell models of cardiovascular health |
Project Leader |
|
Chris Ré |
Stanford |
Intercampus Research Awards |
Multi-scale deep learning and single-cell models of cardiovascular health |
Collaborator |
|
Deepak Srivastava |
UCSF Gladstone Institutes |
Intercampus Research Awards |
Multi-scale deep learning and single-cell models of cardiovascular health |
Collaborator |
|
Tomasz Nowakowski |
UCSF |
Intercampus Research Awards |
Social Network Analysis of neuroimmune interactions in the developing human brain |
Project Leader |
|
Alex Pollen |
UCSF |
Intercampus Research Awards |
Social Network Analysis of neuroimmune interactions in the developing human brain |
Project Leader |
|
David Schaffer |
Berkeley |
Intercampus Research Awards |
Social network analysis of neuroimmune interactions in the developing human brain |
Project Leader |
|
Alice Ting |
Stanford |
Intercampus Research Awards |
Social Network Analysis of neuroimmune interactions in the developing human brain |
Project Leader |
|
Chun Jimmie Ye |
UCSF |
Intercampus Research Awards |
Social Network Analysis of neuroimmune interactions in the developing human brain |
Project Leader |
|
James Zou |
Stanford |
Intercampus Research Awards |
Social Network Analysis of neuroimmune interactions in the developing human brain |
Project Leader |
|
Kole Roybal |
UCSF |
Intercampus Research Awards |
Social Network Analysis of neuroimmune interactions in the developing human brain |
Collaborator |
| James Wells | UCSF | Intercampus Research Awards | Social Network Analysis of neuroimmune interactions in the developing human brain | Collaborator |
APPENDIX 2
INTERCAMPUS RESEARCH AWARDS AND MICROBIOME INITIATIVE TEAM DESCRIPTIONS
Beyond model systems: insights into genome evolution and cellular innovations
Biomedical research usually focuses on a few well-studied model organisms like mice or fruit flies, but some questions in biology are hard to answer using these organisms. For example, it is hard to study limb regeneration in mice, since mice don’t regenerate limbs. The ocean is full of organisms that are built in very different ways and that offer an opportunity to develop new approaches to human health in areas such as regeneration, immunity, stem cell biology, and aging. The team seeks to harness diverse marine organisms in combination with state-of-the-art genomic technology to answer a set of fundamental questions in biology that have gone unanswered for over a century, with a focus on animal development, regeneration, immune function, and genome maintenance and stability.
Defining host responses of virus-infected and uninfected neighbor cells
How viral infections spread within tissues is poorly understood. Infected cells can induce changes in neighboring cells to enhance viral spread (pro-viral responses), or to block infection (antiviral responses). The team brings together expertise in virology, immunology, and RNA biology to define the cellular pathways induced in infected and uninfected neighboring cells and their influence on viral replication. The team will focus on how RNA transmission, alterations in viral and cellular RNA stability and nutritional status can elicit pro- or anti-viral responses in neighbors, drawing on their expertise in culturing 3D human organoids to provide realistic models for the complex interactions that occur between cells in human tissue. Understanding the mechanisms affecting viral spread within tissues will lead toward new treatments for viral infections.
Imaging complex biological machines in action
The team will develop an integrated pipeline for determining the structure and function of exceedingly complex molecular machines in intact cells. They will develop new approaches for specimen preparation, data collection, and image analysis so that a single sample can be observed using several complementary imaging technologies that reveal different features and span a range of magnifications. This will allow the team to determine with nearly atomic resolution the structures of molecular machines composed of a large number of distinct proteins while also determining the orientation and position of the machines within cells, providing previously inaccessible functional insights. They will use their methods to characterize the machinery that Toxoplasma and related parasites, like the one responsible for malaria, use to invade human cells.
Machine learning for interpreting rare genetic variation in comprehensive newborn screening and pharmacogenetics
In California, 500,000 babies are born each year, some of whom have genetic mutations that cause disease or altered responses to medications. Recognizing which genetic variants cause problems is surprisingly difficult, impeding the use of genetic information to inform early intervention or the customization of patient care. The team has drawn together experts in biology, computer science, medicine, and ethics to develop new methods for identifying genetic variants that cause disease, focusing on serious newborn diseases and on gene variants that affect patient responses to medications. The team will collect experimental data and develop innovative machine learning techniques to predict the functional consequences of genetic variants.
Multi-scale deep learning and single-cell models of cardiovascular health
The team will develop methods to accelerate the pace of discovery of genetic determinants for cardiovascular disease. They will develop new statistical machine learning tools to analyze morphological and functional parameters of the heart from clinical images, an approach that can be scaled to analyze millions of images. They will also develop machine learning tools based on enhanced iterative random forests (iRF) to identify genetic variants likely to account for some of the variation in cardiovascular morphology and function observed in their analysis of clinical images, utilizing publicly available large-scale clinical data sets and local patient cohorts. Finally, they will identify genetic variants responsible for functional phenotypes using cell-based in vitro model systems.
Social network analysis of neuroimmune interactions in the developing human brain
The team seeks to develop scalable technologies for studying intercellular interactions in the human brain. They will establish a panel of synthetic antibodies that recognize distinct cell types in the human brain and develop tools (“molecular flight recorders”) to monitor and record the history of single cells as they respond to signals and interact with neighboring cells. The team will apply these tools to understand how the specialized innate immune cells in the brain (microglia) interact with other types of brain cells, interactions that have been proposed to play a key role during normal brain development and in psychiatric and neurodegenerative diseases. The tools and technologies developed by the team will be broadly applicable to study interactions between many cell types in diverse organs and tissues.
Microbiome Initiative
Humans are colonized by communities of microbes that influence many aspects of human health, from nutrition and immune function to drug metabolism. Although technological advances have made It easier to enumerate which microbes are present in different individuals and how the composition of microbial communities changes over time, the mechanisms by which these microbial communities contribute to human biology are not well understood. A major question is how changes in an individual’s microbiome and differences in the composition of the microbiome between individuals influence human health. The team will take advantage of new technologies to interrogate and manipulate the microbiome, approaching the microbiome from the perspectives of engineering, ecology, and genetics and focusing on methods of precise measurement that will enable mechanistic insight.
Stay up-to-date on the latest news, publications, competitions, and stories from CZ Biohub.
Marketing cookies are required to access this form.
 News Releases
News Releases
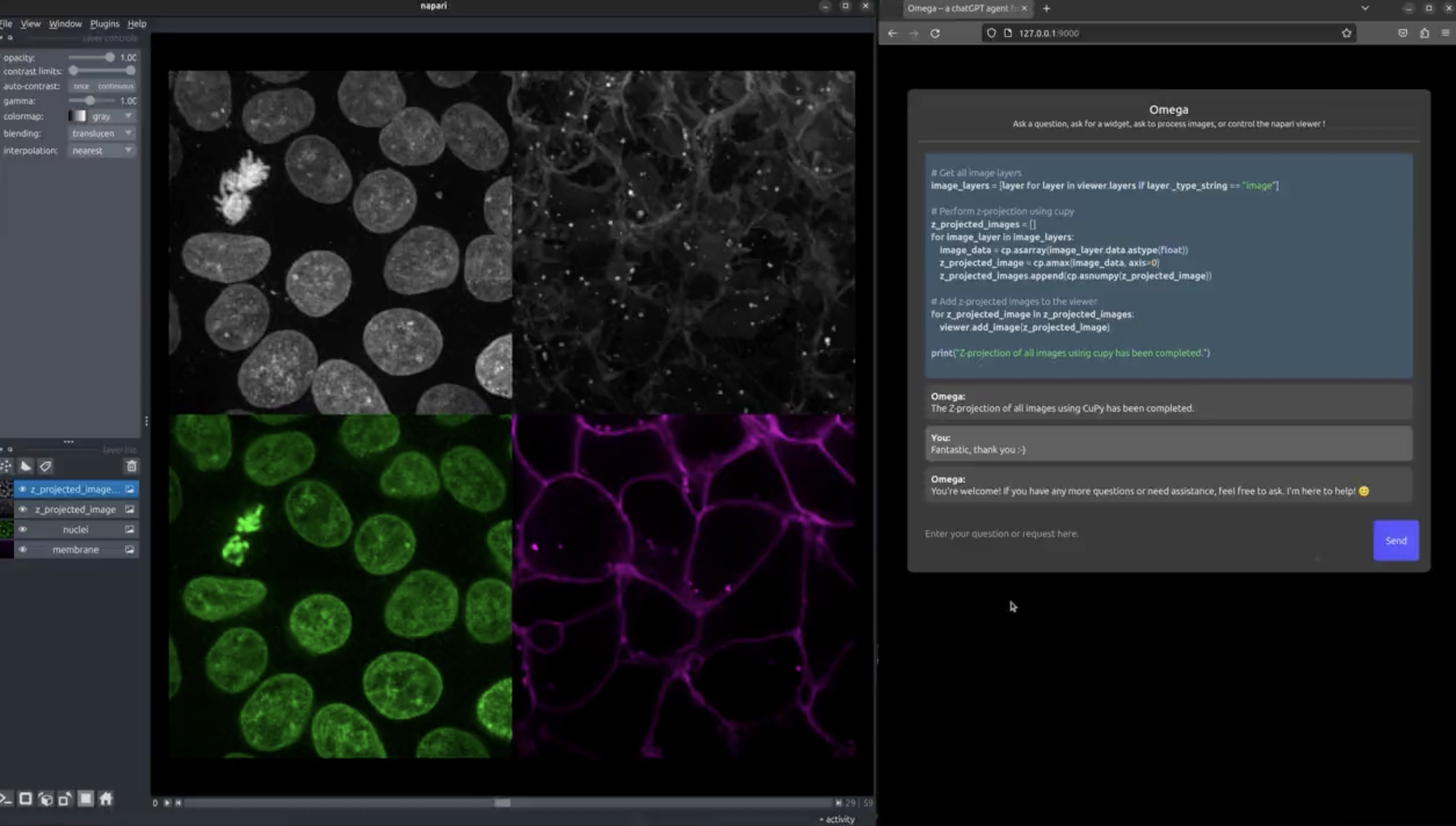
By lowering technical barriers, Omega democratizes image analysis ...
 News Releases
News Releases
Comprehensive mapping of pathogen populations points way to precision treatments to improve ...
 News Releases
News Releases
Join scientist Holger Müller in his lab, which is developing innovative imaging technology that may hold the key to unlocking new insights into our ...
Stay up-to-date on the latest news, publications, competitions, and stories from CZ Biohub.
Marketing cookies are required to access this form.