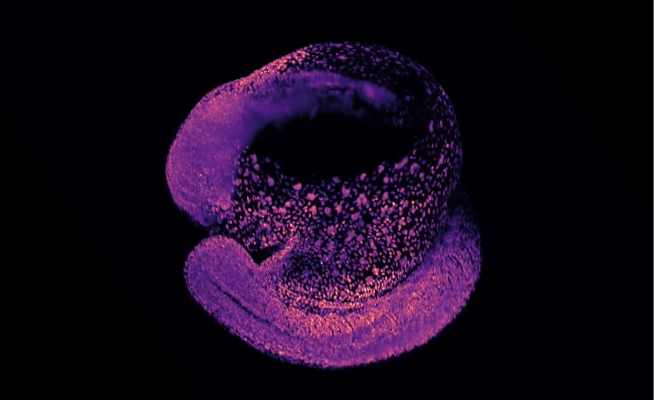
shrimPy is a pythonic framework for correlative 3D imaging of physical and molecular properties of cells at high throughput.
OUR WORK
We create, collaborate, catalyze
We create and openly disseminate knowledge and technologies through collaborative, interdisciplinary research to catalyze science
Cells are the fundamental unit of life. Our goal is to understand dynamic cell systems across scales, in both healthy and diseased states.
We study cell physiology across scales: at the molecular level of transcriptomes, proteomes, and metabolomes; at the whole-cell level of functionally and compositionally distinct organelles; and at the whole-organism level of divergent cell types and changing cell states. And at the population level we collaborate with scientists in low- and middle-income countries, building capacity in state-of-the-art metagenomics and molecular epidemiology to detect and manage infectious diseases.
We develop new technologies for high-throughput, scalable cell and molecular biology; genomics; transcriptomics; customized and automated microscopy; mass spectrometry; and AI and data analytics.
And we build scientific bridges with our partner universities – Stanford University, UC Berkeley, and UCSF – through our Investigator Program, creating and catalyzing connections and a rich culture of scientific exchange. We also partner with institutes across the CZ Biohub Network, created and supported by the Chan Zuckerberg Initiative.
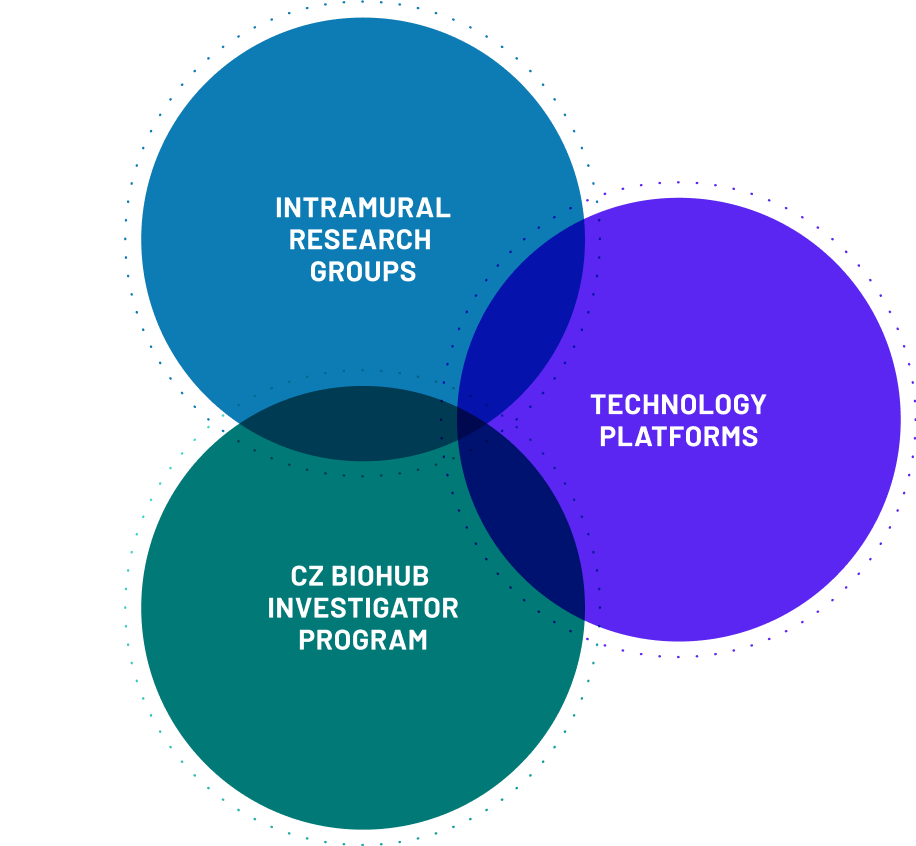

INVESTIGATOR PROGRAM
Building community to catalyze innovation
The Investigator Program at CZ Biohub SF is funding research by world-renowned scientists, engineers, and technologists from Stanford University, UC Berkeley, and UCSF.
Learn moreFeatured Tools & Projects
Subscribe to our bimonthly Nexus newsletter, and receive other updates from CZ Biohub Network.
Cookies and JavaScript are required to access this form.